Machine learning can help read the language of life
DNA is the language of life: our DNA forms a living record of things that went well for our ancestors, and things that didn’t. DNA tells our body (and every other organism) which proteins to produce; these proteins are tiny machines that carry out enormous tasks, from fighting off infection to helping you ace an upcoming exam in school.
But for about a third of all proteins that all organisms produce, we just don’t know what they do. It’s kind of like we’re in a factory where everything’s buzzing, and we’re surrounded by all these impressive tools, but we have only a vague idea of what’s going on. Understanding how these tools operate, and how we can use them, is where we think machine learning can make a big difference.
An example of a previously-solved protein structure (E. coli TrpCF) and the area where our AI makes predictions of its function. This protein produces tryptophan, which is a chemical that's required in your diet to keep your body and brain running.
Recently, DeepMind showed that AlphaFold can predict the shape of protein machinery with unprecedented accuracy. The shape of a protein provides very strong clues as to how the protein machinery can be used, but doesn’t completely solve this question. So we asked ourselves: can we predict what function a protein performs?
In our Nature Biotechnology article, we describe how neural networks can reliably reveal the function of this “dark matter” of the protein universe, outperforming state-of-the-art methods. We worked closely with internationally recognized experts at the EMBL's European Bioinformatics Institute (EMBL-EBI) to annotate 6.8 million more protein regions in the Pfam v34.0 database release, a global repository for protein families and their function. These annotations exceed the expansion of the database over the last decade, and will enable the 2.5 million life-science researchers around the world to discover new antibodies, enzymes, foods, and therapeutics.
The Pfam database is a large collection of protein families and their sequences. Our ML models helped annotate 6.8 million more protein regions in the database.
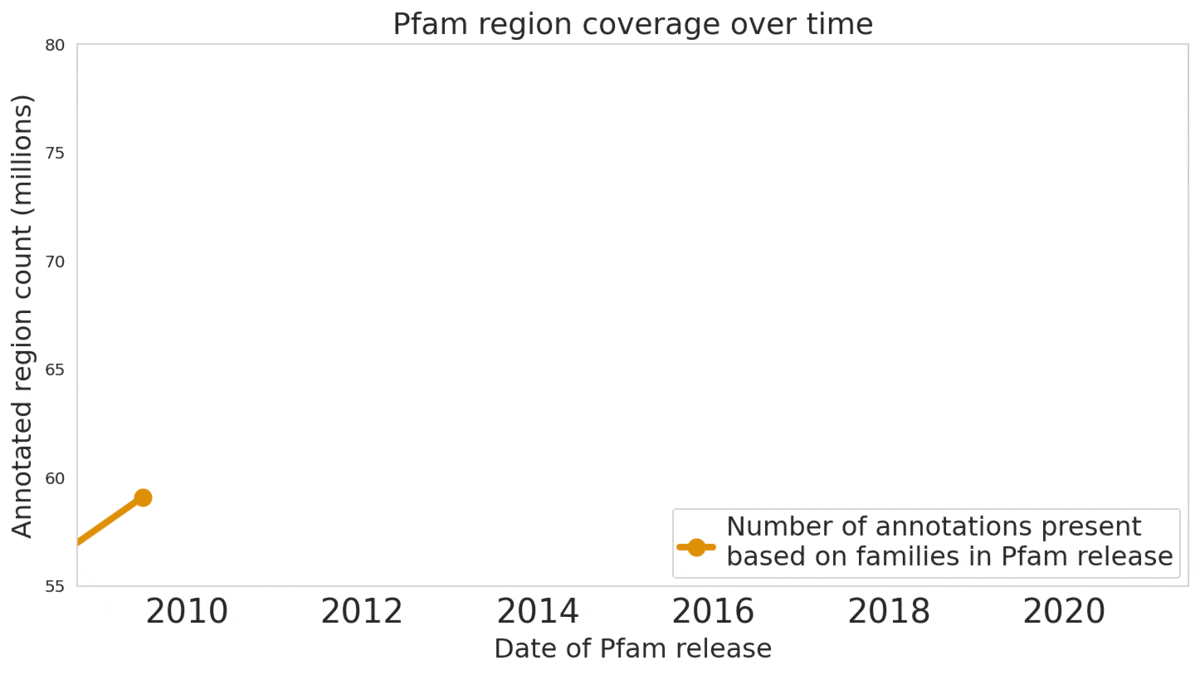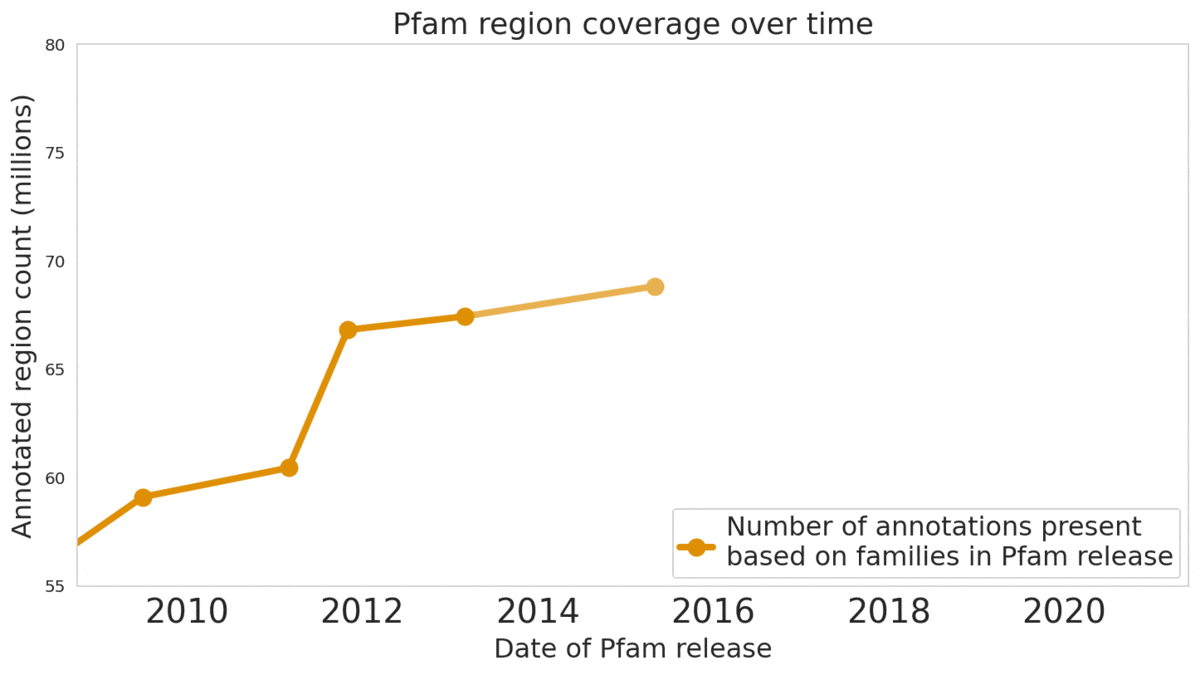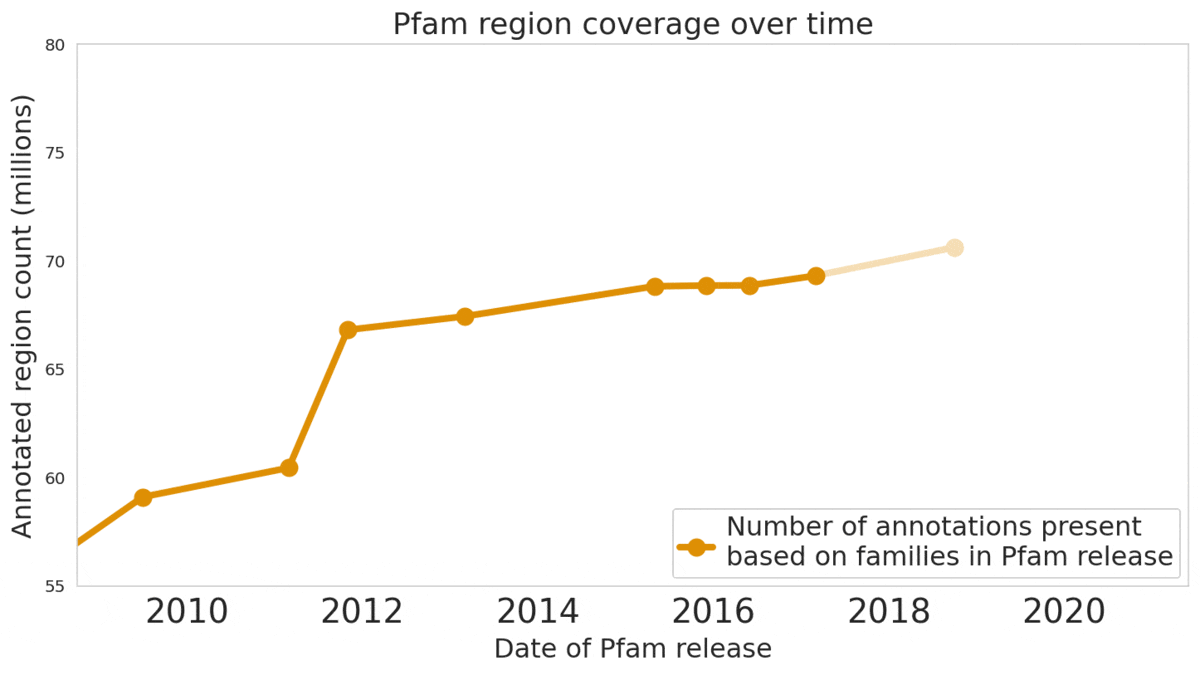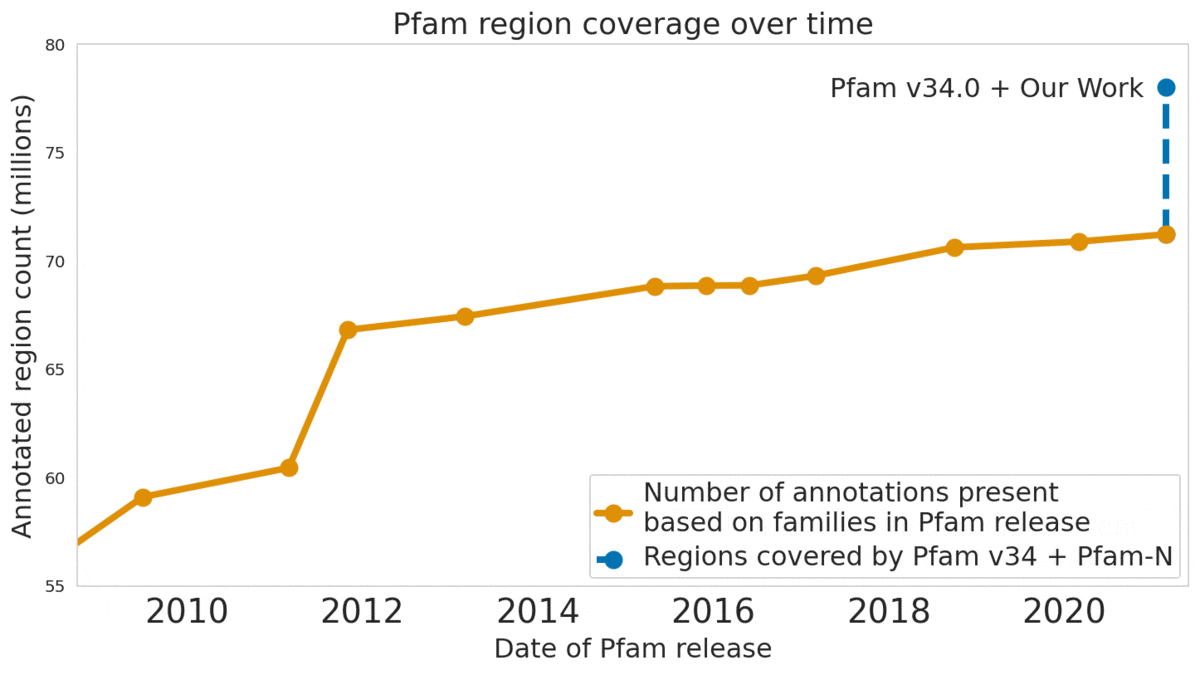
We also understand there’s a reproducibility crisis in science, and we want to be part of the solution — not the problem. To make our research more accessible and useful, we’re excited to launch an interactive scientific article where you can play with our ML models — getting results in real time, all in your web browser, with no setup required.
Google has always set out to help organize the world’s information, and to make it useful to everyone. Equity in access to the appropriate technology and useful instruction for all scientists is an important part of this mission. This is why we’re committed to making these models useful and accessible. Because, who knows, one of these proteins could unlock the solution to antibiotic resistance, and it’s sitting right under our noses.